Chapter 11 Basic Graphics
11.1 Introducing Graphics
Consider the following example:
wt <- mtcars$wt
mpg <- mtcars$mpg
# open a graphics window and generate a scatter plot between automobile weight on the horizontal axis and miles per gallon on the vertical axis
plot(wt, mpg)
# add a line of best fit
abline(lm(mpg~wt))
#add a title
title("Regression of MPG on Weight")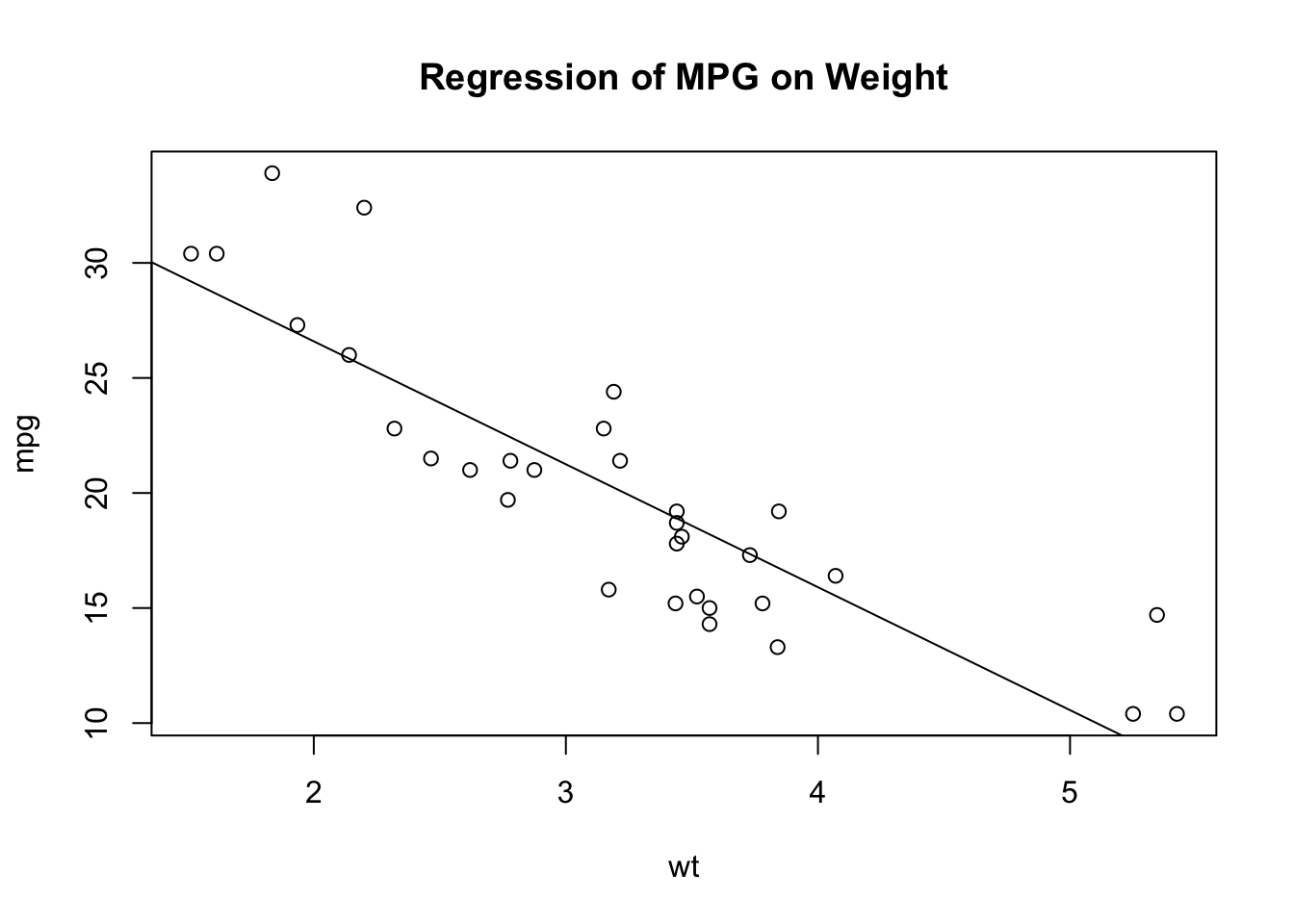
You can save your graphs via code or through GUI menus. To save a graph via code, sandwich the statements that produce the graph between a statement that sets a destination and a statement that closes that destination. For example, the following will save the graph as a PDF document named mygraph.pdf in the current working directory:
pdf("mygraph.pdf")
wt <- mtcars$wt
mpg <- mtcars$mpg
plot(wt, mpg)
abline(lm(mpg~wt))
title("Regression of MPG on Weight")
dev.off()
## quartz_off_screen
## 2In addition to pdf(), you can use the functions win.metafile(), png(), jpeg(), bmp(), tiff(), xfig(), and postscript() to save graphs in other formats.
Saving graphs via the GUI is platform specific. On a Windows platform, select File > Save As from the graphics window, and choose the format and location desired in the resulting dialog. On a Mac, choose File > Save As from the menu bar when the Quartz graphics window is highlighted. The only output format provided is PDF. On a Unix platform, graphs must be saved via code.
11.1.1 Working with Multiple Graphs
Creating a new graph by issuing a high-level plotting command such as plot(), hist() (for histograms), or boxplot() typically overwrites a previous graph. How can you create more than one graph and still have access to each? There are several methods.
First, you can open a new graph window before creating a new graph. In this case, each new graph will appear in the most recently opened window:
dev.new()
statements to create graph 1
dev.new()
statements to create a graph 2 etc.Second, you can access multiple graphs via the GUI. On a Mac platform, you can step through the graphs at any time using Back and Forward on the Quartz menu. On a Windows platform, you must use a two-step process. After opening the first graph window, choose History > Recording. Then use the Previous and Next menu items to step through the graphs that are created.
Finally, you can use the functions dev.new(), dev.next(), dev.prev(), dev.set(), and dev.off() to have multiple graph windows open at one time and choose which output is sent to which windows. This approach works on any platform. See help(dev.cur) for details on this approach.
11.2 Basic Graphical Parameters
We are going to use the following data that describes patient responses to two drugs at five dosage levels:
dose <- c(20, 30, 40, 45, 60)
drugA <- c(16, 20, 27, 40, 60)
drugB <- c(15, 18, 25, 31, 40)A simple line graph relating dose to response for drug A can be created using
plot(dose, drugA, type="b")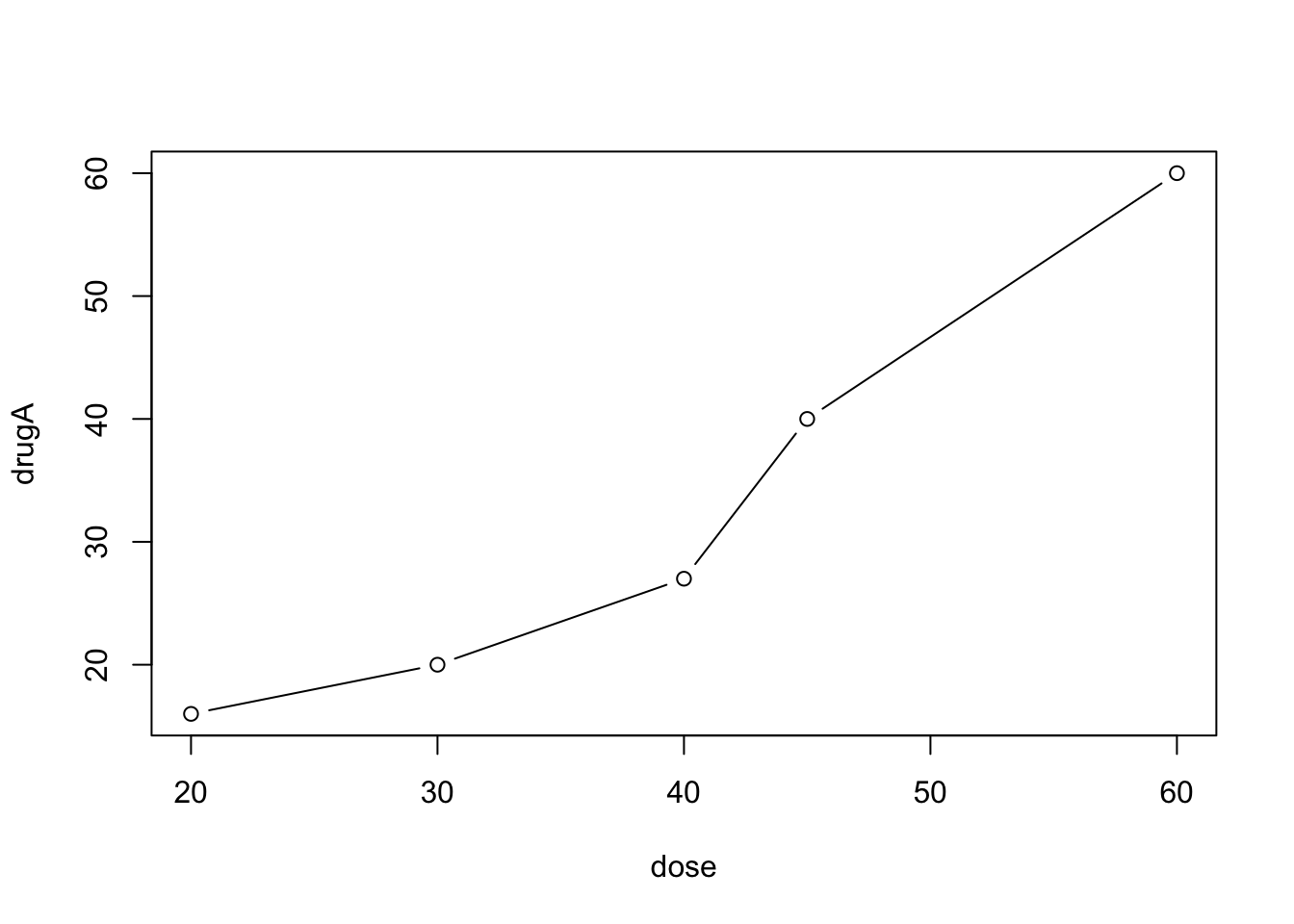 The option
The option type="b" indicates that both points and lines should be plotted. Without type="b", we would get a scatterplot of dose and drugA. Use help(plot) to view other options.
You can customize many features of a graph (fonts, colors, axes, and labels) through options called graphical parameters. One way is to specify these options through the par() function. Values set in this manner will be in effect for the rest of the session or until they’re changed. The format is par(optionname=value, optionname=value, ...). Specifying par() without parameters produces a list of the current graphical settings. Adding the no.readonly=TRUE option produces a list of current graphical settings that can be modified.
Let’s say that you’d like to use a solid triangle rather than an open circle as your plotting symbol, and connect points using a dashed line rather than a solid line. You can do so with the following code:
opar <- par(no.readonly=TRUE) # make a copy of current settings
par(lty=2, pch=17) # change line type to dashed (lty=2) and symbol to triangle (pch=17)
plot(dose, drugA, type="b") # generate a plot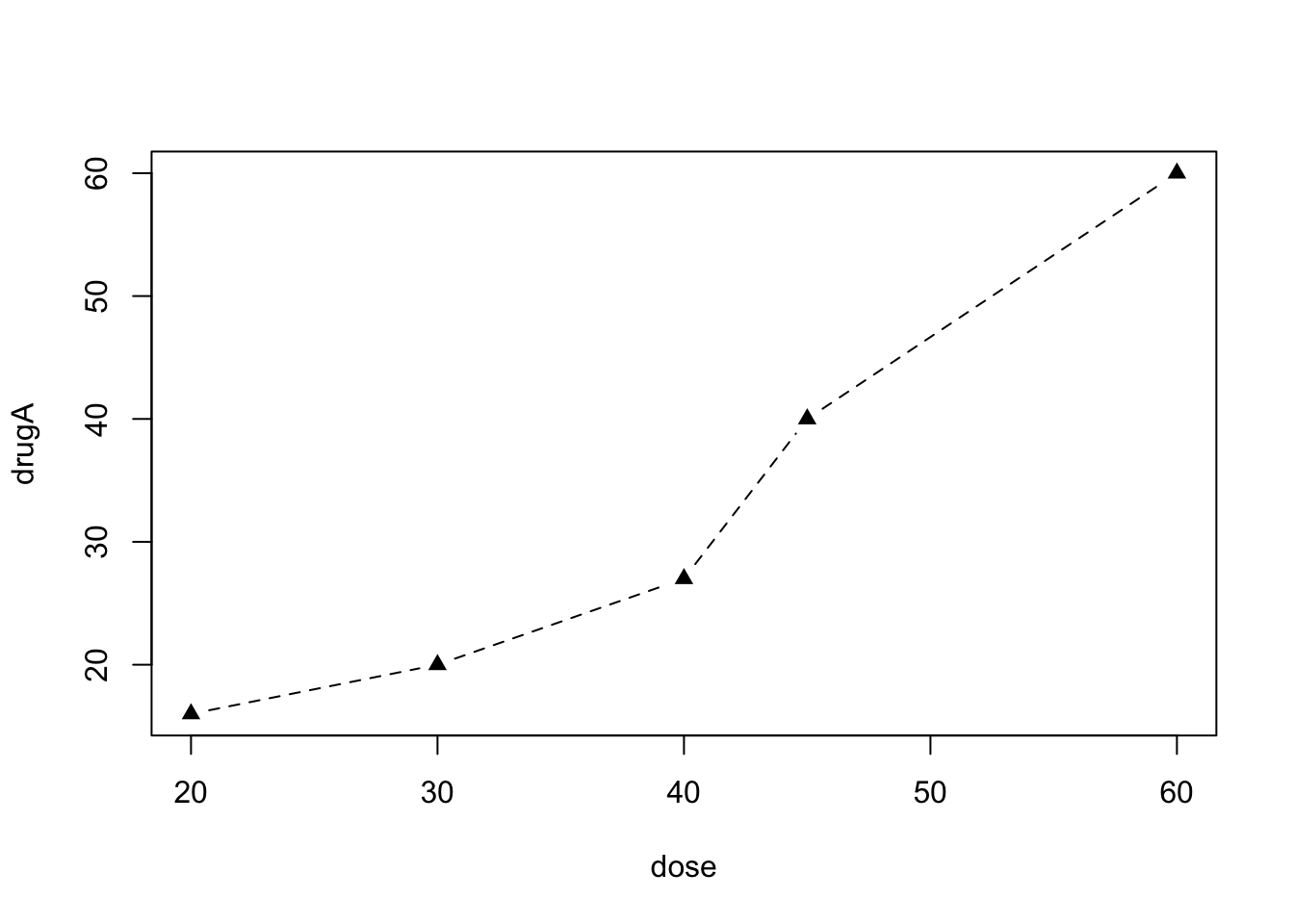
par(opar) # restore the original settings You can have as many par() functions as desired, so par(lty=2, pch=17) could also be written as
par(lty=2)
par(pch=17)A second way to specify graphical parameters is by providing the optionname=value pairs directly to a high-level plotting function. In this case, the options are only in effect for that specific graph. You could generate the same graph with this code:
plot(dose, drugA, type="b", lty=3, lwd=3, pch=17, cex=2)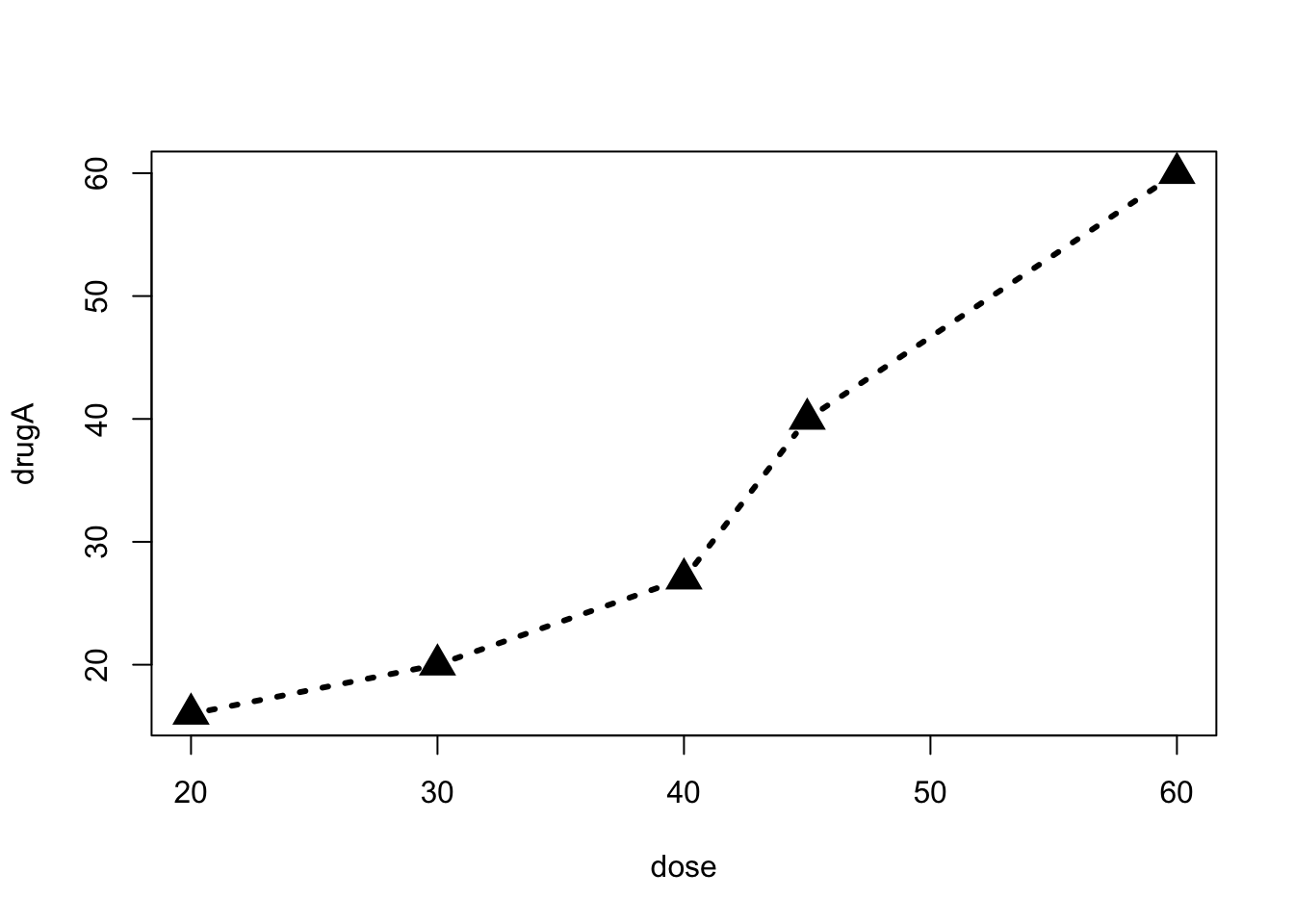 Use
Use help(par) to see the list of gaphical parameters. In the rest of this section, we are basically see some of the most frequently used ones.
11.2.1 Lines and Symbols
pch: Either an integer (between 0 and 25) specifying a symbol or a single character to be used as the default in plotting points. Usehelp(points)to see possible values and their interpretation.cex: A numerical value giving the amount by which plotting text and symbols should be magnified relative to the default. This starts as 1 when a device is opened, and is reset when the layout is changed, e.g. by settingmfrow. There are alsocex.axis,cex.lab,cex.mainthat can be used magnify axis annotation, labels and main titles, respectively.lty: The line type. Line types can either be specified as an integer (0=blank, 1=solid (default), 2=dashed, 3=dotted, 4=dotdash, 5=longdash, 6=twodash) or as one of the character strings “blank”, “solid”, “dashed”, “dotted”, “dotdash”, “longdash”, or “twodash”, where “blank” uses ‘invisible lines’ (i.e., does not draw them).lwd: The line width, a positive number, defaulting to 1.
plot(dose, drugA, type="b", pch = 19, cex=2, lty=3, lwd=3)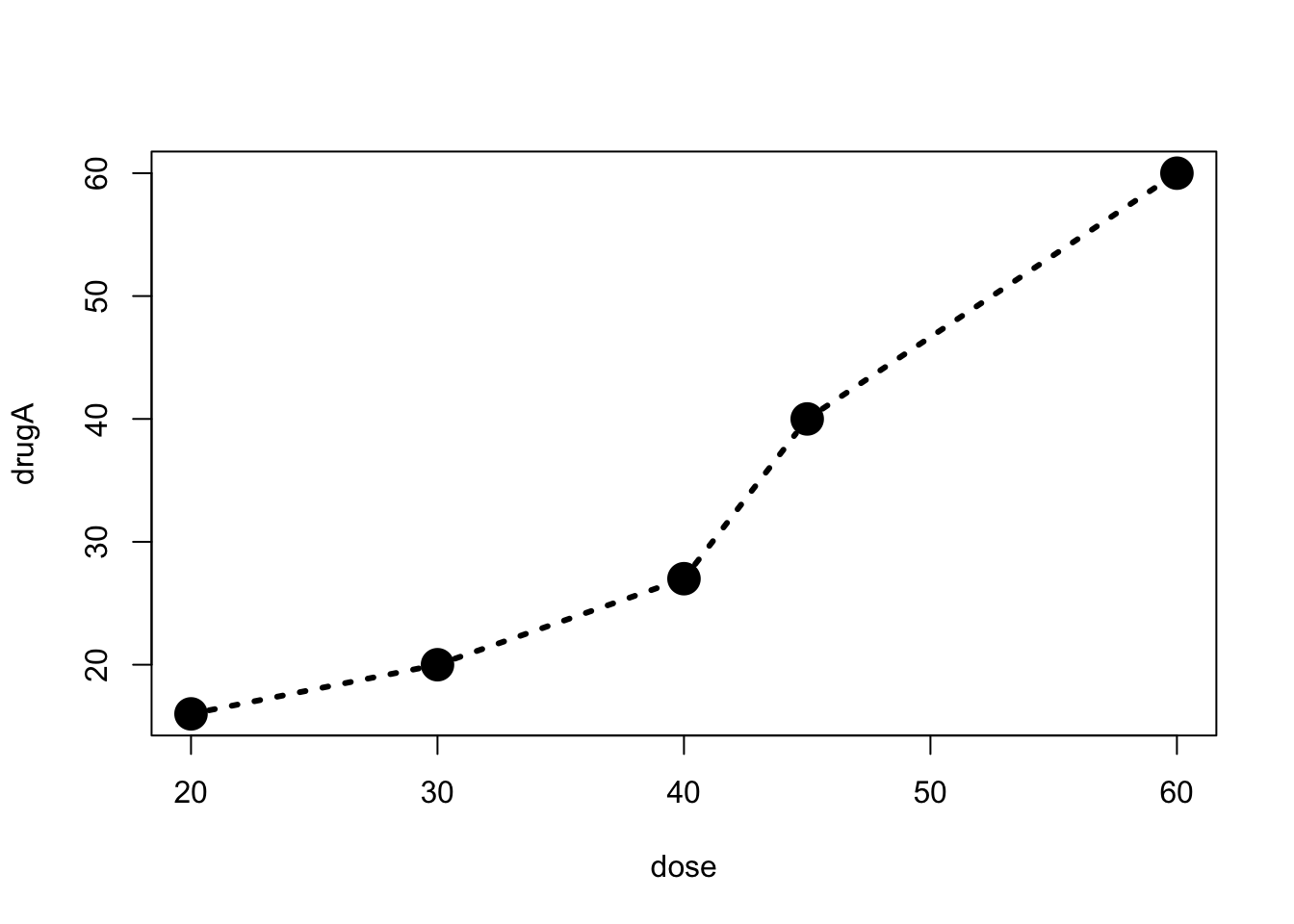 or
or
opar <- par(no.readonly=TRUE) # make a copy of current settings
par(pch = 19, cex=2, lty=3, lwd=3) # set the parameter values
plot(dose, drugA, type="b") # generate a plot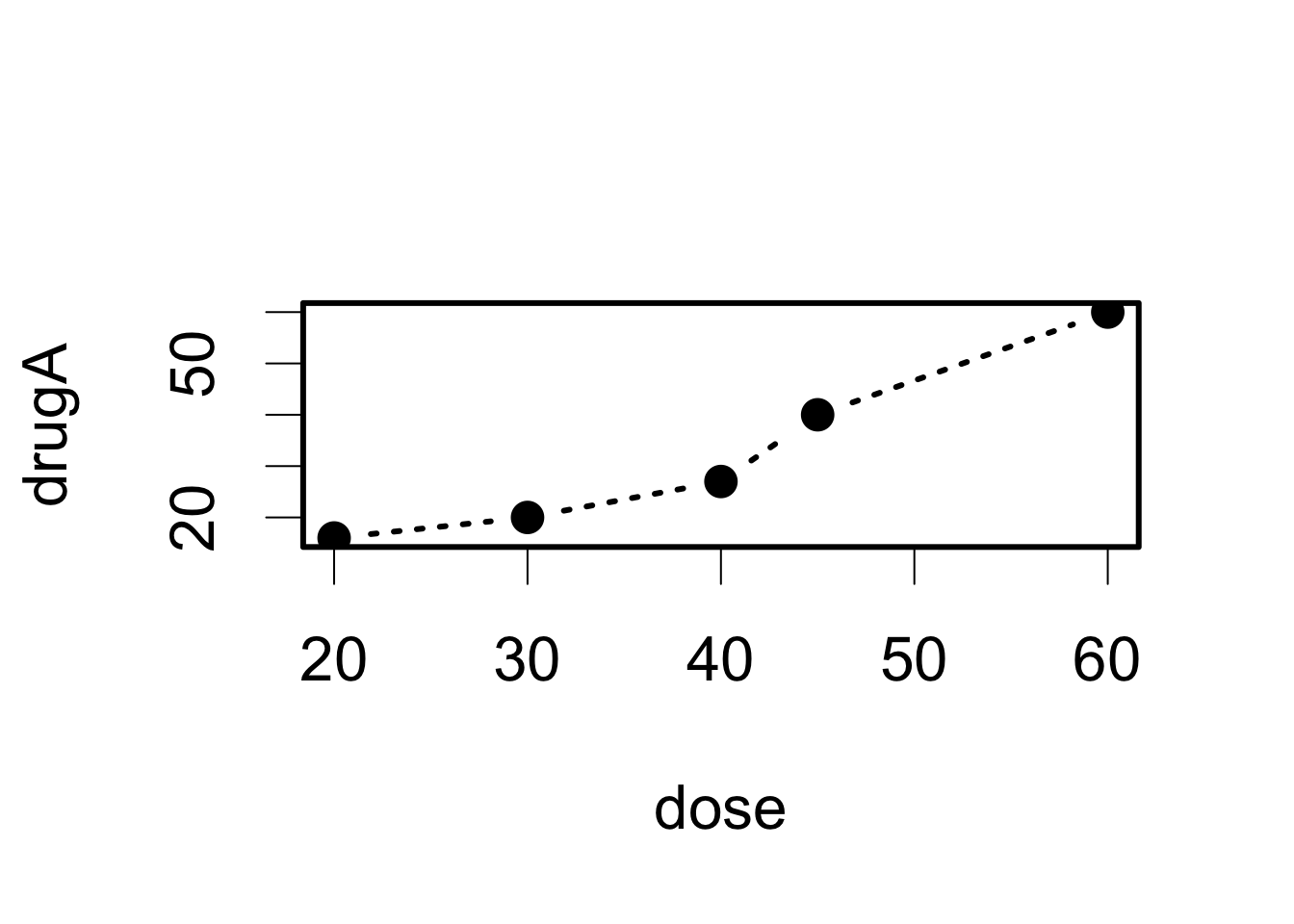
par(opar) # restore the original settings But note that these two appear very differently!
11.2.2 Colors
Some color-related parameters are :
col: A specification for the default plotting color. Colors can be specified in several different ways. The simplest way is with a character string giving the color name (e.g., “red”). A list of the possible colors can be obtained with the functioncolors(). Alternatively, colors can be specified directly in terms of their RGB components with a string of the form"#RRGGBB"where each of the pairs RR, GG, BB consist of two hexadecimal digits giving a value in the range 00 to FF. The functionsrgb,hsv,hcl,grayandrainbowprovide additional ways of generating colors.Some functions (such as
linesandpie) accept a vector of values that are recycled. For example, ifcol=c("red", "blue")and three lines are plotted, the first line will be red, the second blue, and the third red.There are also
col.axis,col.lab,col.main, andcol.subthat can be used to specify colors of axis, axis labels, main titles and subtitles.fg: The color to be used for the foreground of plots. This is the default color used for things like axes and boxes around plots. When called frompar()this also sets parametercolto the same value.bg: The color to be used for the background of the device region."transparent": is transparent, useful for filled areas (such as the background!), and just invisible for things like lines or text. Semi-transparent colors are available for use on devices that support them.gray: Creates a vector of colors from a vector of gray levels. You need to specify gray levels as a vector of numbers between 0 and 1. For example,
plot(dose, drugA, type="b", pch = 19, cex=2, lty=3, lwd=3, col="violet", col.axis="#008000", col.lab=rgb(.5,.1,.4))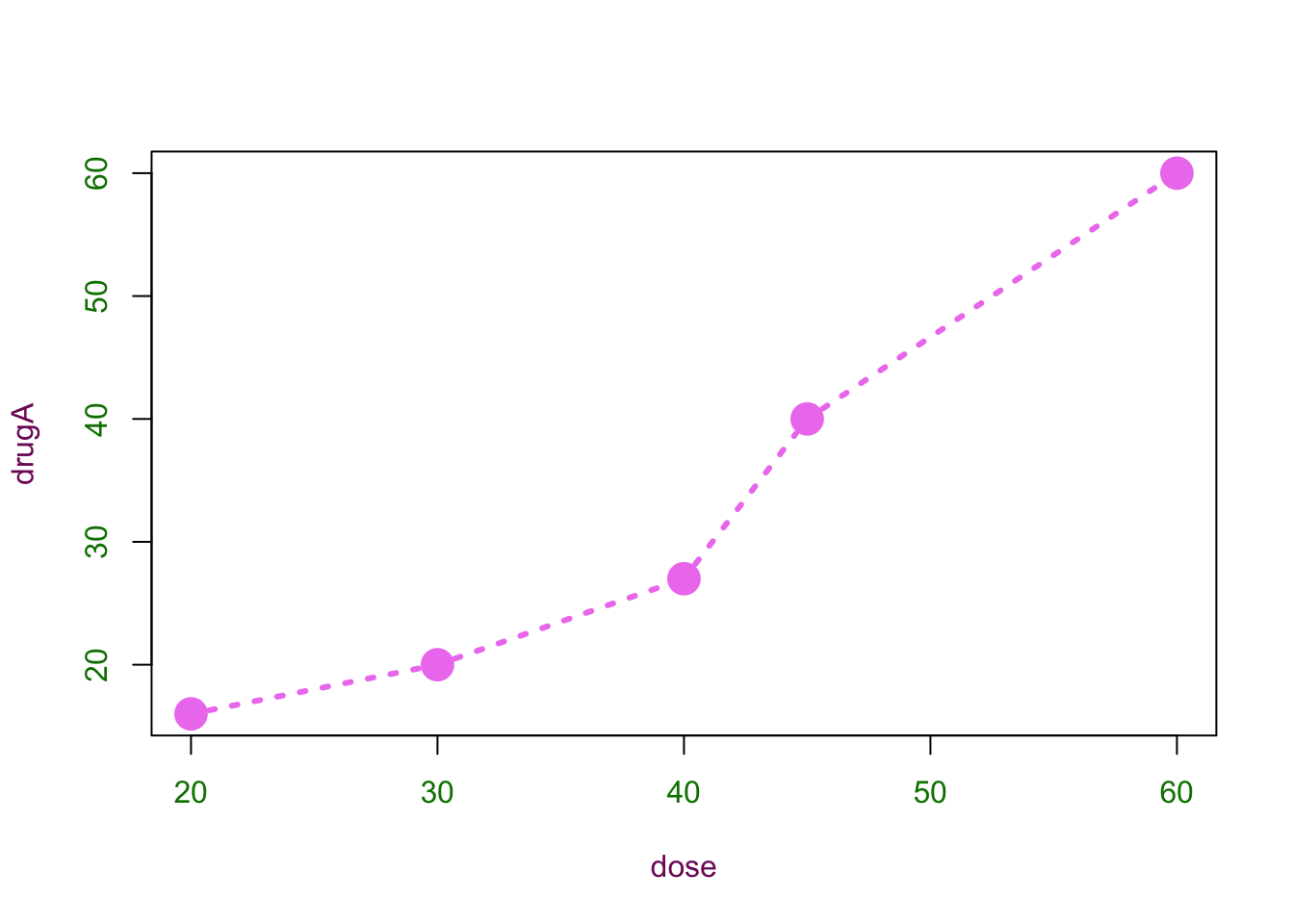
mygrays <- gray(0:4/4)
pie(rep(1, 8), labels=mygrays, col=mygrays)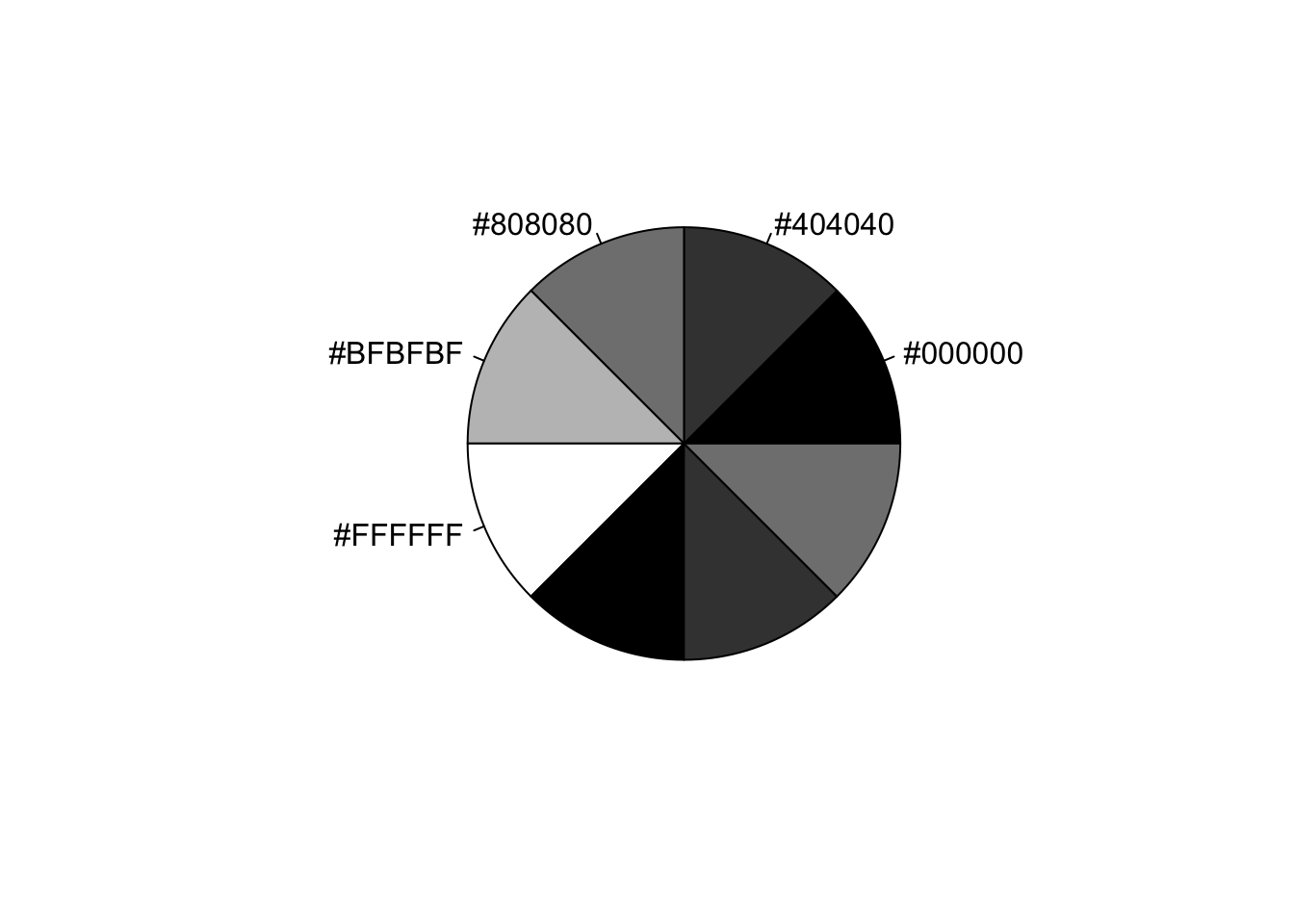
11.2.3 Text Appearance
Graphic parameters that can be used to specify text size, font, and style.
cex: A numerical value giving the amount by which plotting text and symbols should be magnified relative to the default. This starts as 1 when a device is opened, and is reset when the layout is changed, e.g. by settingmfrow. There are alsocex.axis,cex.lab,cex.mainandcex.subthat can be used magnify axis annotation, labels, main titles, and subtitles, respectively.font: vice drivers arrange so that 1 corresponds to plain text (the default), 2 to bold face, 3 to italic and 4 to bold italic. Also, font 5 is expected to be the symbol font, in Adobe symbol encoding. There are alsofont.axis,font.lab,font.mainandfont.subthat can be used for axis annotation, labels, main titles, and subtitles, respectively.ps: Font point size (roughly 1/72 inch). The text size = ps*cex.family: The name of a font family for drawing text. Standard values areserif,sans, andmono.
opar <- par(no.readonly=TRUE) # make a copy of current settings
par(font.lab=2, ps=15) # set the parameter values
plot(dose, drugA, type="b") # generate a plot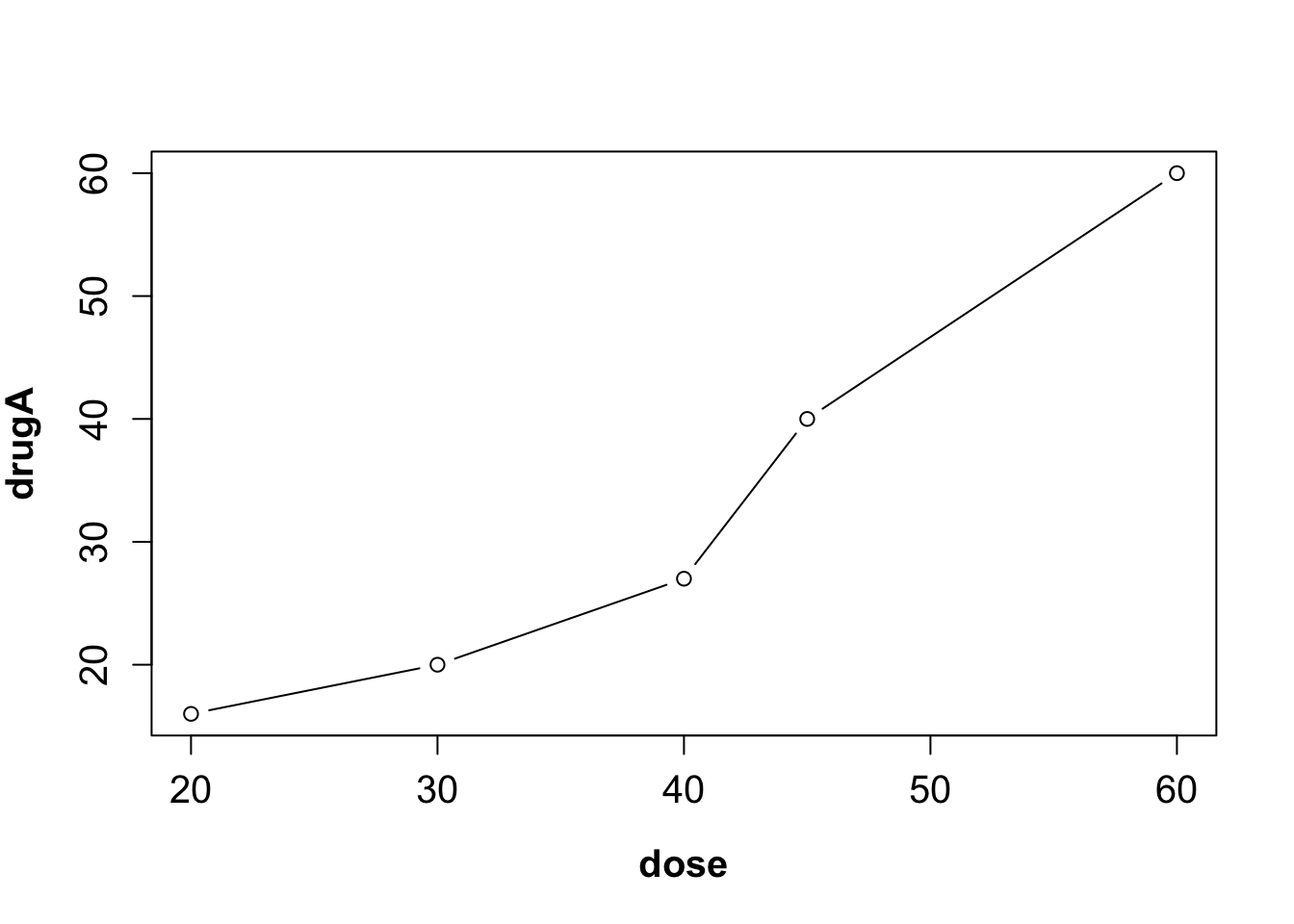
par(opar) # restore the original settings If graphs will be output in PDF format, use names(pdfFonts()) to find out which fonts are available on your system and pdf(file="myplot.pdf", family="fontname") to generate the plots.
11.2.4 Graph Dimensions and Margins
pin: The current plot dimensions, (width, height), in inches.
mai: A numerical vector of the form c(bottom, left, top, right) which gives the margin size specified in inches.
mar: A numerical vector of the form c(bottom, left, top, right) which gives the number of lines of margin to be specified on the four sides of the plot. The default is c(5, 4, 4, 2) + 0.1.
opar <- par(no.readonly=TRUE)
par(pin=c(2,4), mai=c(1,1.5, 1, 0))
plot(dose, drugA, type="b") 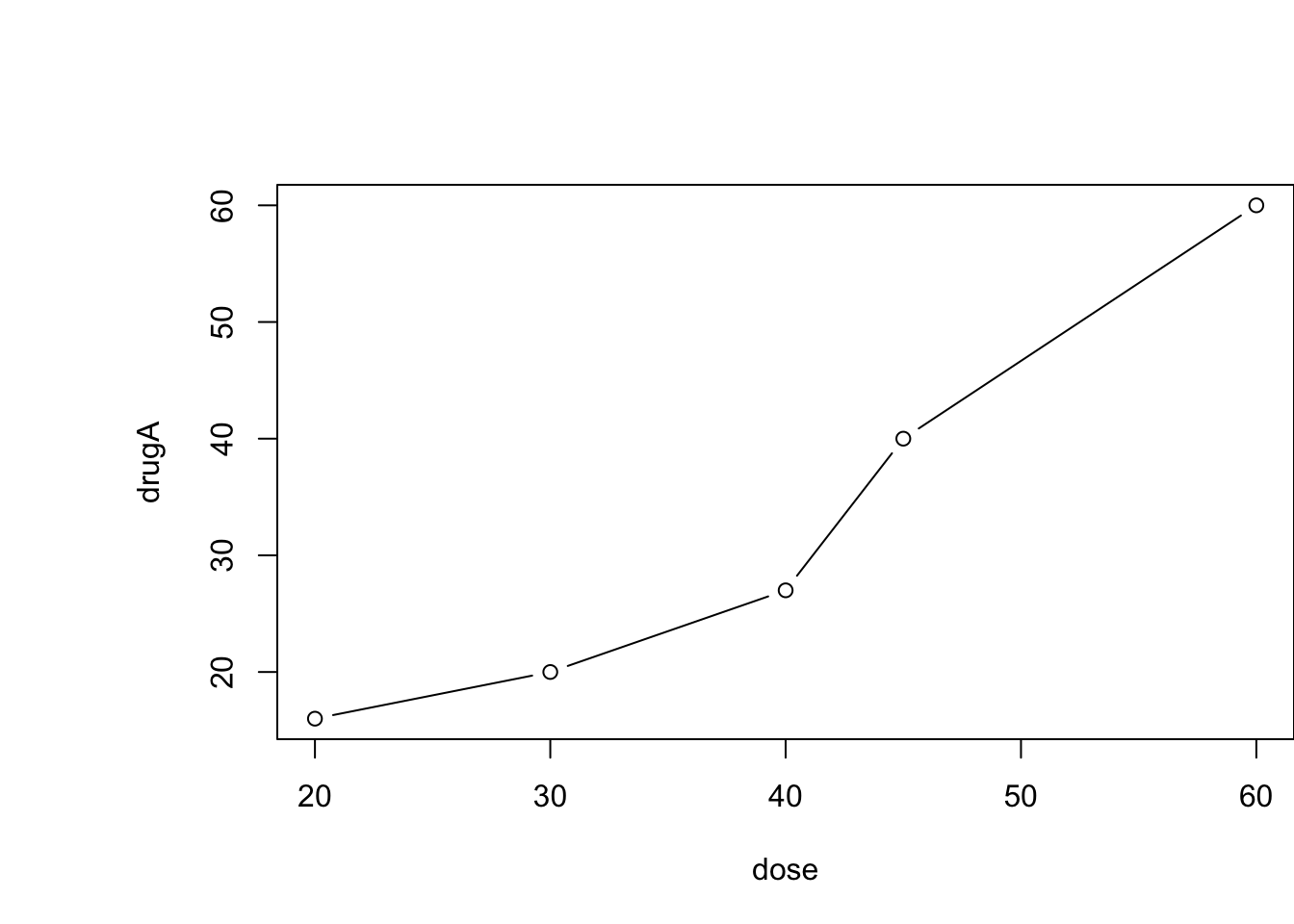
par(opar) Another example:
opar <- par(no.readonly=TRUE)
par(pin=c(3, 2)) # graphs will be 3 inches wide by 2 inches tall
par(lwd=2.0, cex=1.5) # lines will be twice and symbols will be 1.5 times the default
par(cex.axis=.75, font.axis=3) # axis text is set to italic and scaled to 75% of the default.
plot(dose, drugA, type="b", pch=19, lty=2, col="red")
plot(dose, drugB, type="b", pch=23, lty=6, col="blue", bg="green") 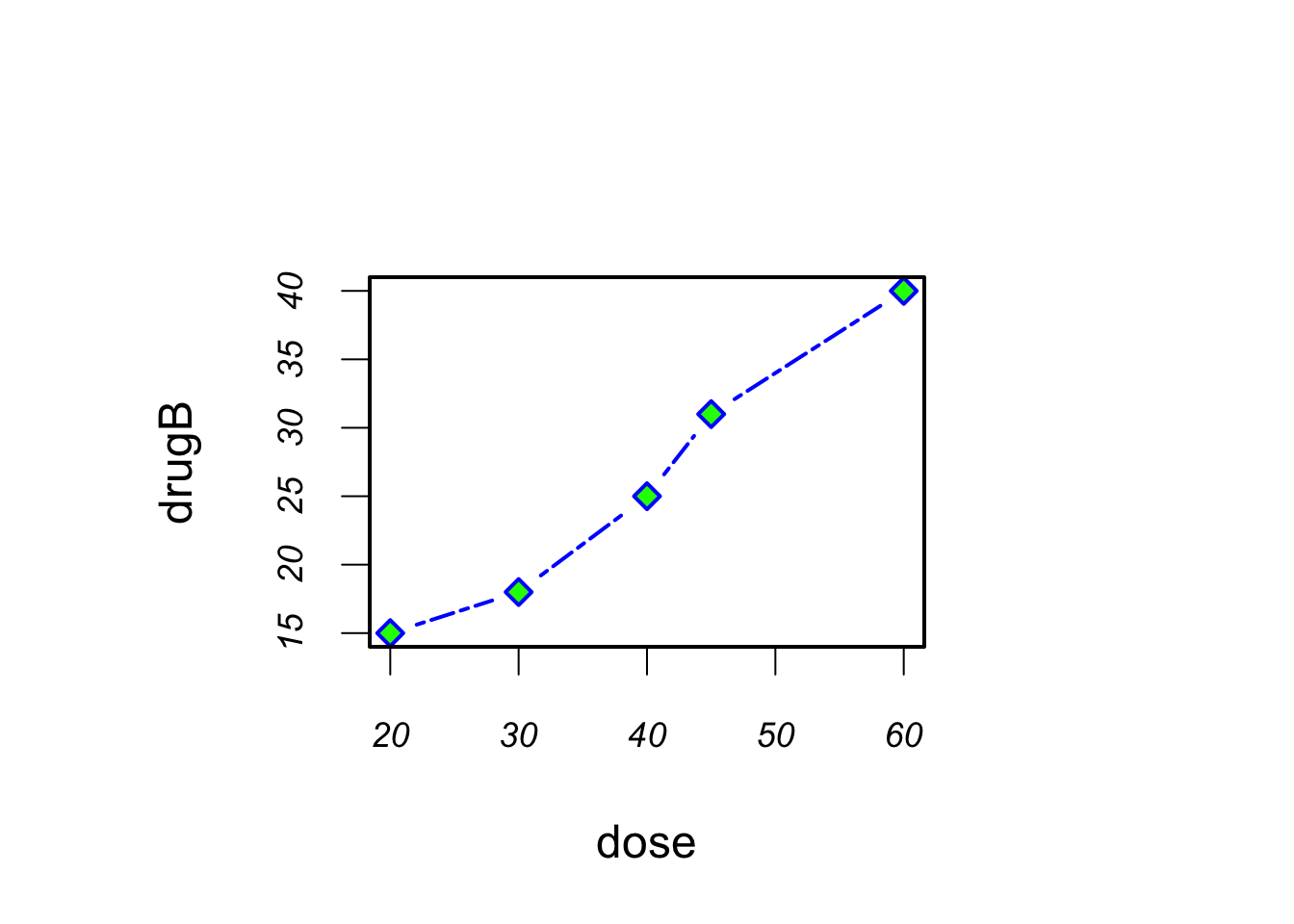
par(opar) Note that parameters set with the par() function apply to both graphs, whereas parameters specified in the plot() functions only apply to that specific graph.
11.2.5 Titles, Axes, Legends, Annotations
Some of the high-level plotting functions (for example, plot, hist, and boxplot) allow you to include axis and text options, as well as graphical parameters:
plot(dose, drugA, type="b",
col="red", lty=2, pch=2, lwd=2,
main="Clinical Trials for Drug A",
sub="This is hypothetical data",
xlab="Dosage", ylab="Drug Response",
xlim=c(0, 60), ylim=c(0, 70))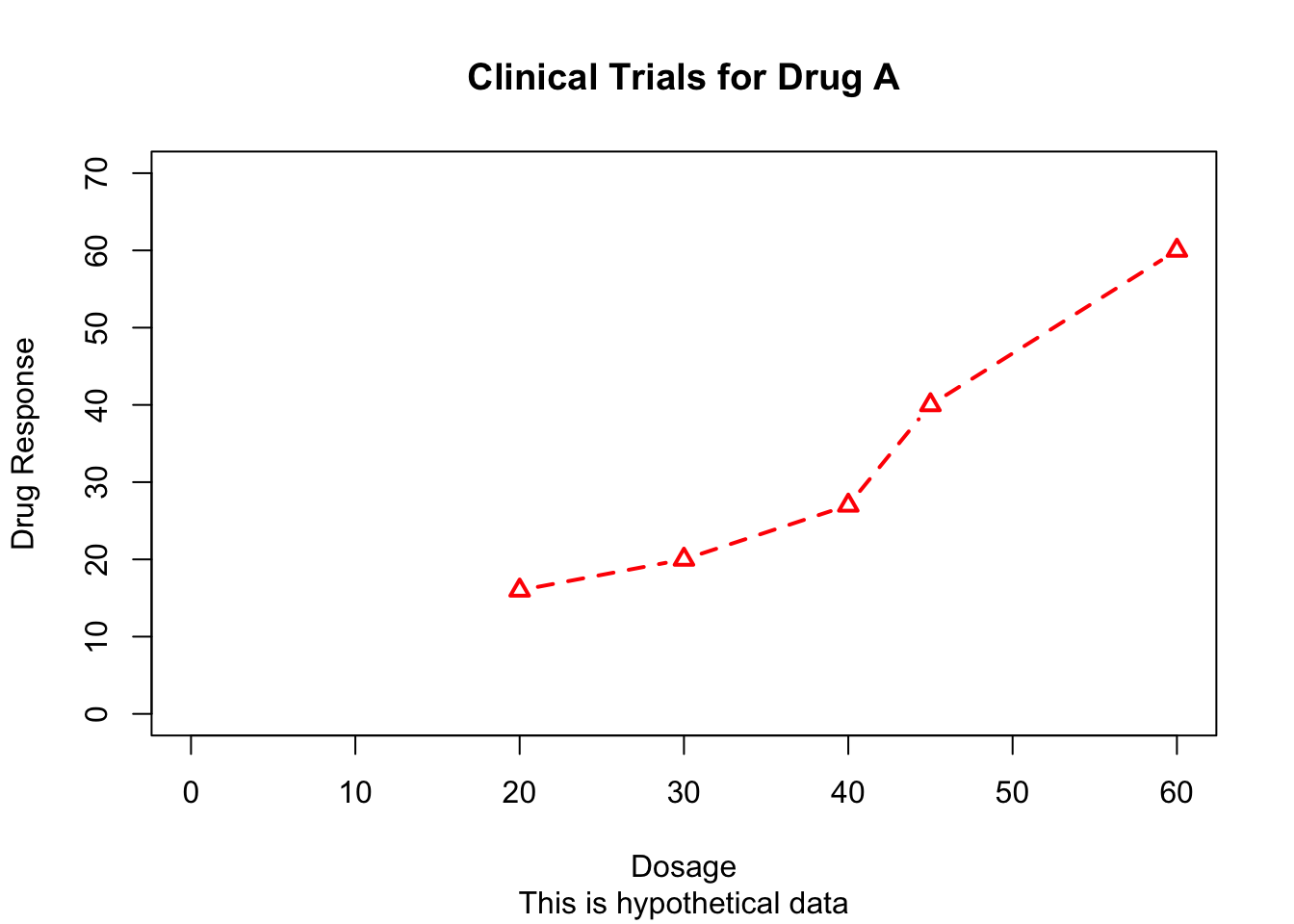
11.2.5.1 title()
This function can be used to add labels to a plot. Its first four principal arguments can also be used as arguments in most high-level plotting functions. The general format is:
title(main="main title", sub="subtitle",
xlab="x-axis label", ylab="y-axis label")Some high-level plotting functions include default titles and labels. You can remove them by adding ann=FALSE in the plot() statement or in a separate par() statement.
Graphical parameters (such as text size, font, rotation, and color) can also be specified in title(). For example, the following code produces a red title and a blue subtitle, and creates green x and y labels that are 25% smaller than the default text size:
title(main="My Title", col.main="red",
sub="My Subtitle", col.sub="blue",
xlab="My X label", ylab="My Y label",
col.lab="green", cex.lab=0.75)axis(): Rather than use R’s default axes, you can create custom axes with the axis() function. The format isaxis(side, at=, labels=, pos=, lty=, col=, las=, tck=, ...)where
side: Integer indicating the side of the graph on which to draw the axis (1 = bottom, 2 = left, 3 = top, and 4 = right).at: Numeric vector indicating where tick marks should be drawn.labels: Character vector of labels to be placed at the tick marks (ifNULL, the at values are used).pos: Coordinate at which the axis line is to be drawn (that is, the value on the other axis where it crosses).col: Line type.las: Line and tick mark color.tck: Specifies that labels are parallel (= 0) or perpendicular (= 2) to the axis. Length of each tick mark as a fraction of the plotting region (a negative number is outside the graph, a positive number is inside, 0 suppresses ticks, and 1 creates gridlines). The default is –0.01.(...): other graphical parameters may also be passed as arguments to this function, particularly, cex.axis, col.axis and font.axis for axis annotation, mgp and xaxp or yaxp for positioning, tck or tcl for tick mark length and direction, las for vertical/horizontal label orientation, or fg instead of col, and xpd for clipping.
When creating a custom axis, you should suppress the axis that’s automatically generated by the high-level plotting function. The option axes=FALSE suppresses all axes (including all axis frame lines, unless you add the option frame.plot=TRUE). The options xaxt="n" and yaxt="n" suppress the x-axis and y-axis, respectively (leaving the frame lines, without ticks).
An example of custom axes:
# Specify data
x <- c(1:10)
y <- x
z <- 10/x
opar <- par(no.readonly=TRUE)
# increase margins
par(mar=c(5, 4, 4, 8) + 0.1)
# plots x vs. y, suppressing annotations
plot(x, y, type="b",
pch=21, col="red",
yaxt="n", lty=3, ann=FALSE)
# add an x versus 1/x line
lines(x, z, type="b", pch=22, col="blue", lty=2)
# draw the axes
axis(2, at=x, labels=x, col.axis="red", las=2)
axis(4, at=z, labels=round(z, digits=2),
col.axis="blue", las=2, cex.axis=0.7, tck=-.01)
# add titles and text
mtext("y=1/x", side=4, line=3, cex.lab=1, las=2, col="blue")
title("An Example of Creative Axes",
xlab="X values",
ylab="Y=X")
par(opar) A plot() statement starts a new graph. By using line() instead, you can add new graph elements to an existing graph. The mtext() function is used to add text to the margins of the plot.
11.2.5.2 Reference lines
The abline() function is used to add reference lines to a graph. The format is
abline(h=yvalues, v=xvalues)Other graphical parameters (such as line type, color, and width) can also be specified in the abline() function. For example
abline(h=c(1,5,7))adds solid horizontal lines at y = 1, 5, and 7, whereas the code
abline(v=seq(1, 10, 2), lty=2, col="blue")adds dashed blue vertical lines at x = 1, 3, 5, 7, and 9.
11.2.5.3 Legend
When more than one set of data or group is incorporated into a graph, a legend can help you to identify what’s being represented by each bar, pie slice, or line. A legend can be added with the legend() function. The format is
legend(location, title, legend, ...)where
location: There are several ways to indicate the location of the legend. You can give an x,y coordinate for its upper-left corner. You can uselocator(1), in which case you use the mouse to indicate the legend’s location. You can also use the keyword bottom, bottomleft, left, topleft, top, topright, right, bottomright, or center to place the legend in the graph. If you use one of these keywords, you can also useinset=to specify an amount to move the legend into the graph (as a fraction of the plot region).title: Character string for the legend title (optional).legend: Character vector with the labels....: Other options. If the legend labels colored lines, specifycol=and a vector of colors. If the legend labels point symbols, specifypch=and a vector of point symbols. If the legend labels line width or line style, uselwd=orlty=and a vector of widths or styles. To create colored boxes for the legend (common in bar, box, and pie charts), usefill=and a vector of colors.Other common legend options include
btyfor box type,bgfor background color,cexfor size, andtext.colfor text color. Specifyinghoriz=TRUEsets the legend horizontally rather than vertically. For more on legends, seehelp(legend). The examples in the help file are particularly informative.
Let’s take a look at an example using the drug data:
dose <- c(20, 30, 40, 45, 60)
drugA <- c(16, 20, 27, 40, 60)
drugB <- c(15, 18, 25, 31, 40)
opar <- par(no.readonly=TRUE)
# Increases line, text, symbol, and label size
par(lwd=2, cex=1.5, font.lab=2)
# Generate the graph
plot(dose, drugA, type="b",
pch=15, lty=1, col="red", ylim=c(0, 60),
main="Drug A vs. Drug B",
xlab="Drug Dosage", ylab="Drug Response")
lines(dose, drugB, type="b",
pch=17, lty=2, col="blue")
abline(h=c(30), lwd=1.5, lty=2, col="gray")
# add minor tick marks
# install.packages("Hmisc") # if not already installed
library(Hmisc)
## Loading required package: lattice
## Loading required package: Formula
##
## Attaching package: 'Hmisc'
## The following objects are masked from 'package:base':
##
## format.pval, units
minor.tick(nx=3, ny=3, tick.ratio=0.5)
# add a legend
legend("topleft", inset=.05, title="Drug Type", c("A","B"), lty=c(1, 2), pch=c(15, 17), col=c("red", "blue"))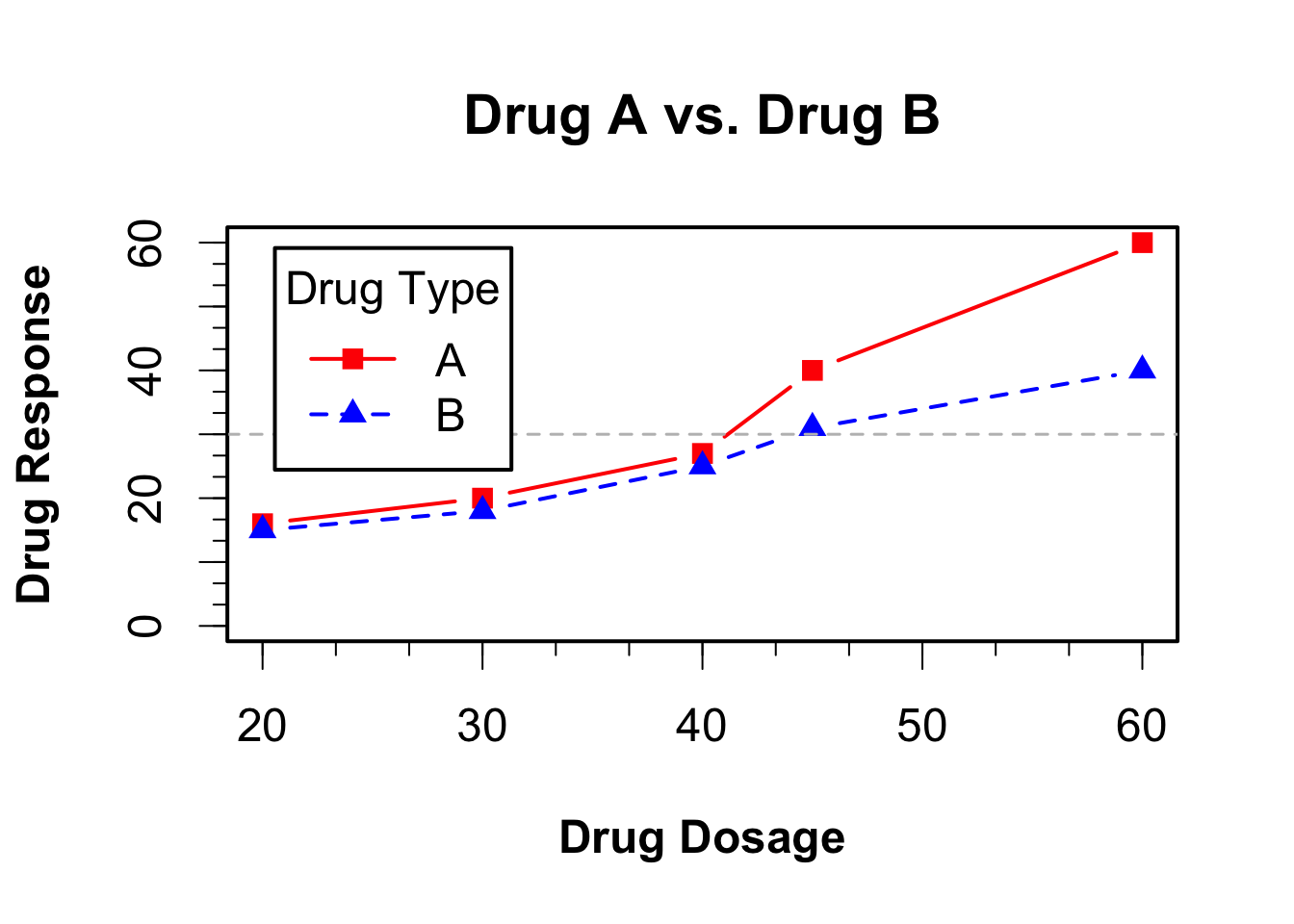
par(opar)11.2.5.4 Text Annotations
Text can be added to graphs using the text() and mtext() functions. text() places text within the graph, whereas mtext() places text in one of the four margins. The formats are
text(location, "text to place", pos, ...)
mtext("text to place", side, line=n, ...)and the common options are:
location: Location can be an x,y coordinate. Alternatively, you can place the text interactively via mouse by specifying location aslocator(1).pos: Position relative to location. 1 = below, 2 = left, 3 = above, and 4 = right. If you specifypos, you can specifyoffset=as a percentage of character width.side: Which margin to place text in, where 1 = bottom, 2 = left, 3 = top, and 4 = right. You can specifyline=to indicate the line in the margin, starting with 0 (closest to the plot area) and moving out. You can also specifyadj=0for left/bottom align- ment oradj=1for top/right alignment.Other common options are
cex,col, andfont(for size, color, and font style, respectively).
The text() function is typically used for labeling points as well as for adding other text annotations. Specify location as a set of x,y coordinates, and specify the text to place as a vector of labels. The x, y, and label vectors should all be the same length. An example is given next:
plot(mtcars$wt, mtcars$mpg,
main="Mileage vs. Car Weight",
xlab="Weight", ylab="Mileage",
pch=18, col="blue")
text(mtcars$wt, mtcars$mpg,
row.names(mtcars),
cex=0.6, pos=4, col="red")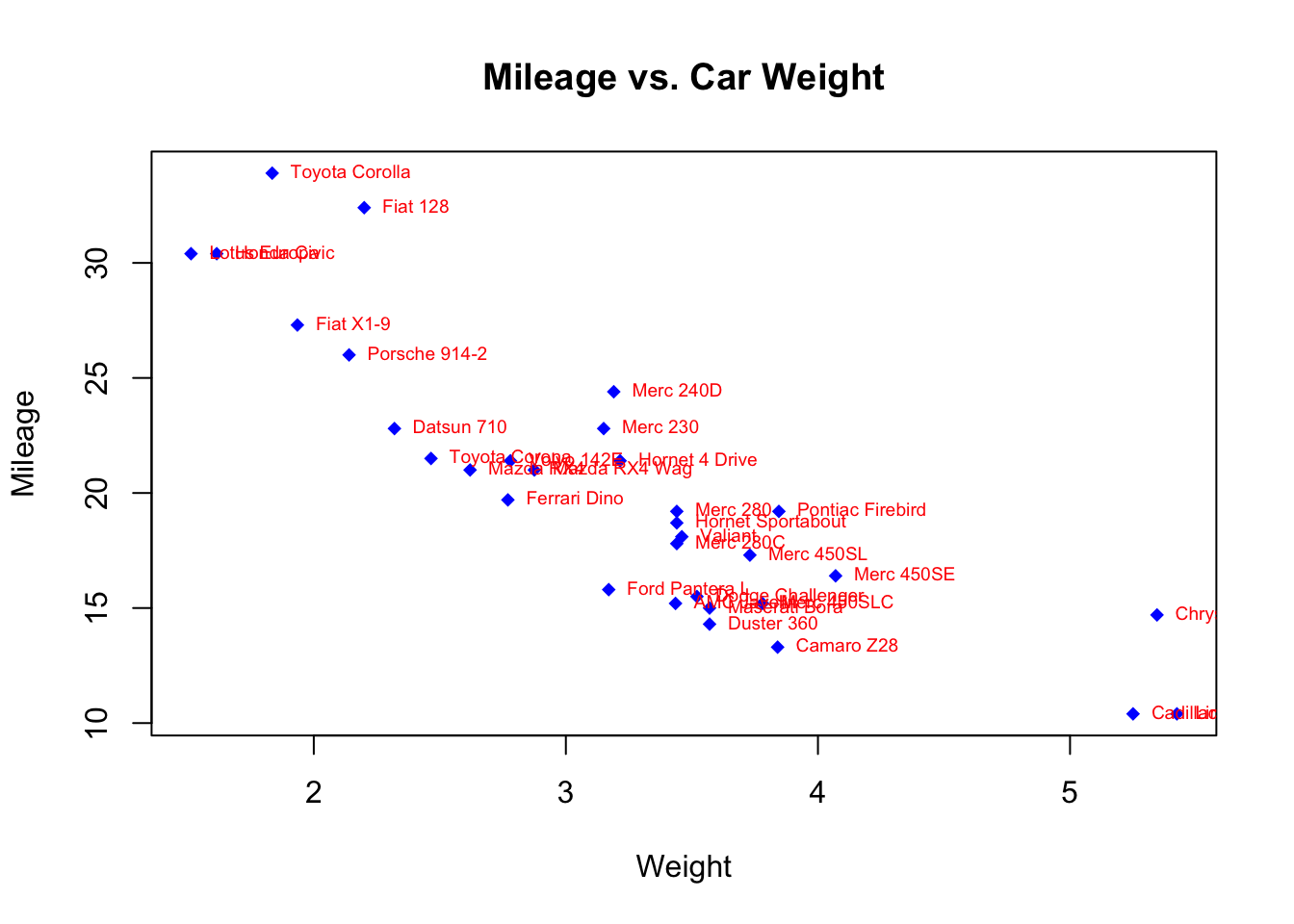
This example plots car mileage versus car weight for the 32 automobile makes provided in the mtcars data frame. The text() function is used to add the car make to the right of each data point. The point labels are shrunk by 40% and presented in red.
As a second example, the following code can be used to display font families:
opar <- par(no.readonly=TRUE)
par(cex=1.5)
plot(1:7,1:7,type="n")
text(3,3,"Example of default text")
text(4,4,family="mono","Example of mono-spaced text")
text(5,5,family="serif","Example of serif text")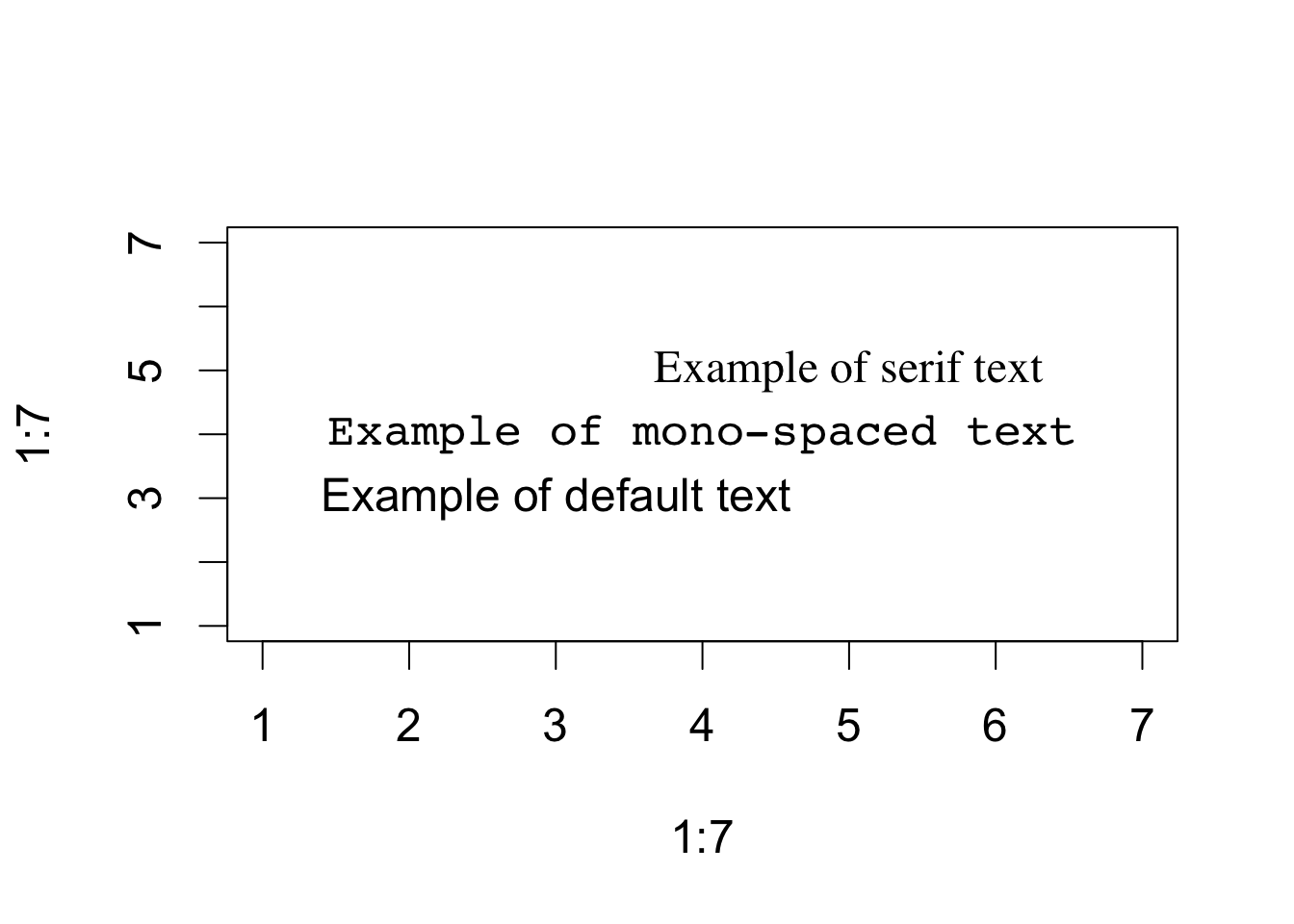
par(opar) The resulting plot will differ from platform to platform, because plain, mono, and serif text are mapped to different font families on different systems.
11.2.5.5 Math annotations
Finally, you can add mathematical symbols and formulas to a graph using TeX-like rules. See help(plotmath) for details and examples. You can also try demo(plotmath) to see this in action. The plotmath() function can be used to add mathematical symbols to titles, axis labels, or text annotations in the body or margins of a graph.
11.3 Combining Graphs: par(), layout()
R makes it easy to combine several graphs into one overall graph, using either the par() or layout() function.
With the par() function, you can include the graphical parameter mfrow=c(nrows, ncols) to create a matrix of nrows × ncols plots that are filled in by row. Alternatively, you can use mfcol=c(nrows, ncols) to fill the matrix by columns.
For example, the following code creates four plots and arranges them into two rows and two columns:
# Combining graphs
opar <- par(no.readonly=TRUE)
par(mfrow=c(2,2))
plot(mtcars$wt, mtcars$mpg, main="Scatterplot of wt vs. mpg")
plot(mtcars$wt, mtcars$disp, main="Scatterplot of wt vs. disp")
hist(mtcars$wt, main="Histogram of wt")
boxplot(mtcars$wt, main="Boxplot of wt")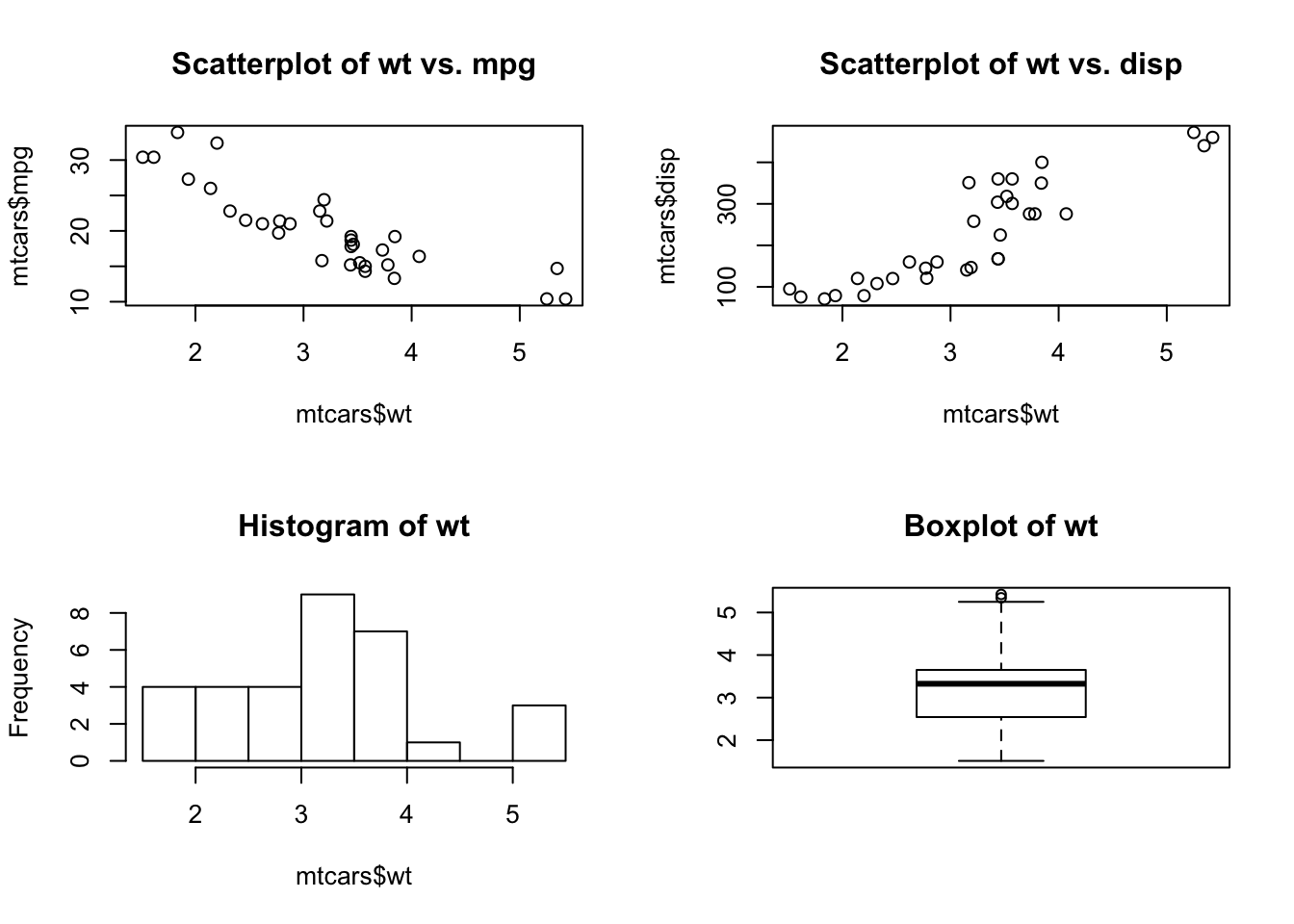
par(opar)As a second example, let’s arrange three plots in three rows and one column:
opar <- par(no.readonly=TRUE)
par(mfrow=c(3,1))
hist(mtcars$wt)
hist(mtcars$mpg)
hist(mtcars$disp)
par(opar)Note that the high-level function hist() includes a default title (use main="" to suppress it, or ann=FALSE to suppress all titles and labels).
The layout() function has the form layout(mat), where mat is a matrix object specifying the location of the multiple plots to combine. In the following code, one figure is placed in row 1 and two figures are placed in row 2:
layout(matrix(c(1,1,2,3), 2, 2, byrow = TRUE))
hist(mtcars$wt)
hist(mtcars$mpg)
hist(mtcars$disp)
Optionally, you can include widths= and heights= options in the layout() function to control the size of each figure more precisely. widths is a vector of values for the widths of columns and heights is a vector of values for the heights of rows.
Relative widths are specified with numeric values. Absolute widths (in centimeters) are specified with the lcm() function.
In the following code, one figure is again placed in row 1 and two figures are placed in row 2. But the figure in row 1 is one-third the height of the figures in row 2. Additionally, the figure in the bottom-right cell is one-fourth the width of the figure in the bottom-left cell:
layout(matrix(c(1, 1, 2, 3), 2, 2, byrow = TRUE),
widths=c(1, 0.5), heights=c(0.5, 1))
hist(mtcars$wt)
hist(mtcars$mpg)
hist(mtcars$disp)As you can see, layout() gives you easy control over both the number and place- ment of graphs in a final image and the relative sizes of these graphs. See help(layout) for more details.
There are times when you want to arrange or superimpose several figures to create a single meaningful plot. Doing so requires fine control over the placement of the figures. You can accomplish this with the fig= graphical parameter. In the following example, two box plots are added to a scatter plot to create a single enhanced graph:
opar <- par(no.readonly=TRUE)
par(fig=c(0, 0.8, 0, 0.8))
# sets up the scatter plot
plot(mtcars$wt, mtcars$mpg,
xlab="Miles Per Gallon",
ylab="Car Weight")
# adds a box plot above
par(fig=c(0, 0.8, 0.55, 1), new=TRUE)
boxplot(mtcars$wt, horizontal=TRUE, axes=FALSE)
# adds a box plot to the right
par(fig=c(0.65, 1, 0, 0.8), new=TRUE)
boxplot(mtcars$mpg, axes=FALSE)
# add title and set up other parameters
mtext("Enhanced Scatterplot", side=3, outer=TRUE, line=-3)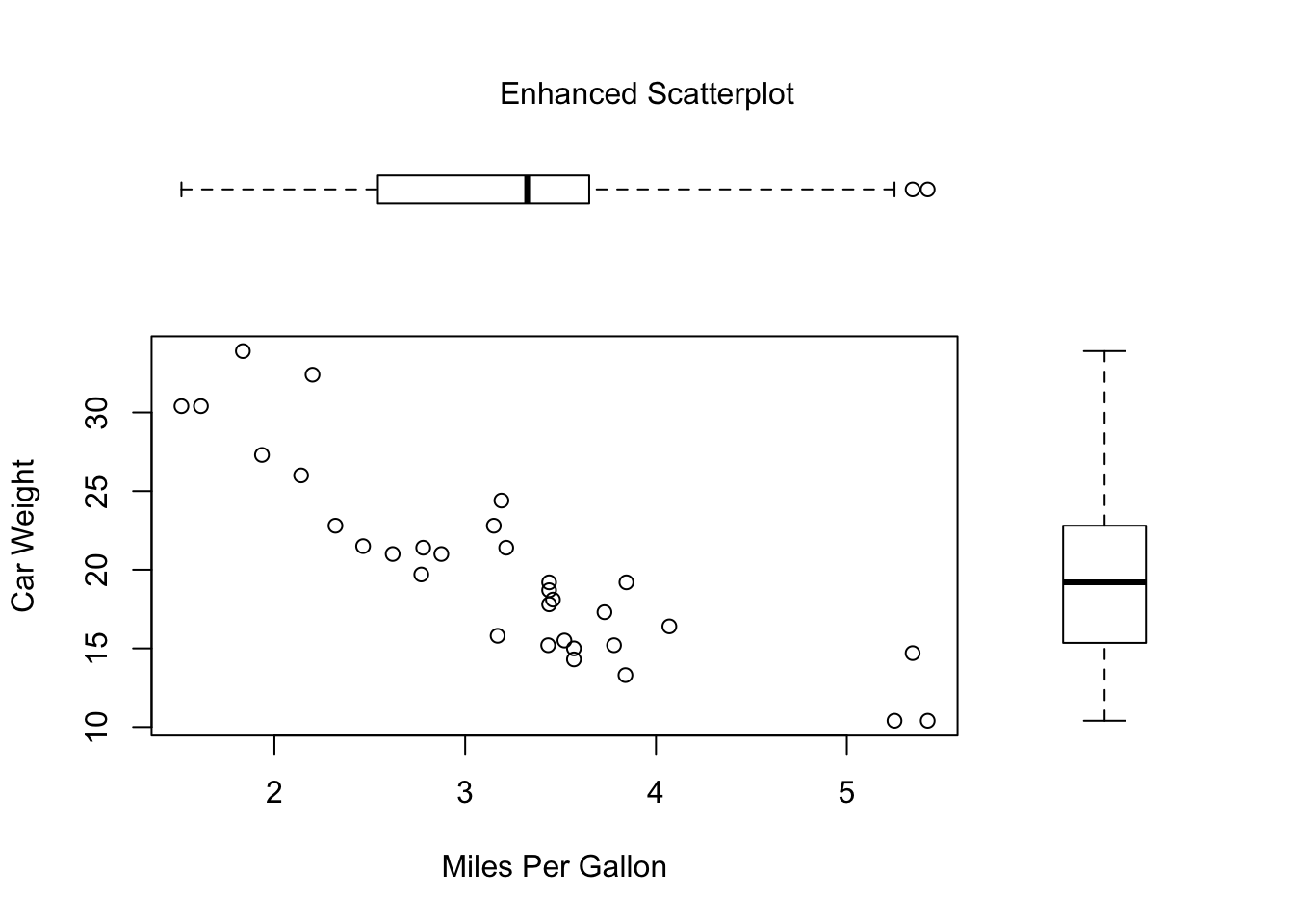
par(opar)o understand how this graph is created, think of the full graph area as going from (0,0) in the lower-left corner to (1,1) in the upper-right corner. The format of the fig= parameter is a numerical vector of the form c(x1, x2, y1, y2).
The first fig= sets up the scatter plot going from 0 to 0.8 on the x-axis and 0 to 0.8 on the y-axis. The top box plot goes from 0 to 0.8 on the x-axis and 0.55 to 1 on the y-axis. The box plot on the right goes from 0.65 to 1 on the x-axis and 0 to 0.8 on the y-axis. fig= starts a new plot, so when you add a figure to an existing graph, include the new=TRUE option.
I chose 0.55 rather than 0.8 so that the top figure would be pulled closer to the scatter plot. Similarly, I chose 0.65 to pull the box plot on the right closer to the scatter plot. You have to experiment to get the placement correct.
The amount of space needed for individual subplots can be device dependent. If you get “Error in plot.new(): figure margins too large,” try varying the area given for each portion of the overall graph.
You can use the fig= graphical parameter to combine several plots into any arrangement within a single graph. With a little practice, this approach gives you a great deal of flexibility when creating complex visual presentations.